C9orf72 ALS shows muted microglial response
ALS driven by the C9orf72 hexanucleotide repeat expansion shows a fundamentally different glial response than sporadic ALS, despite near-identical clinical presentation. The findings by the team of Renzo Mancuso at the VIB-UAntwerp Center for Molecular Neurology, together with Philip Van Damme at KU Leuven/UZ Leuven, and the Van Den Bosch lab at the VIB-KU Leuven Center for Biology of Disease Research, were published in Nature Neuroscience.
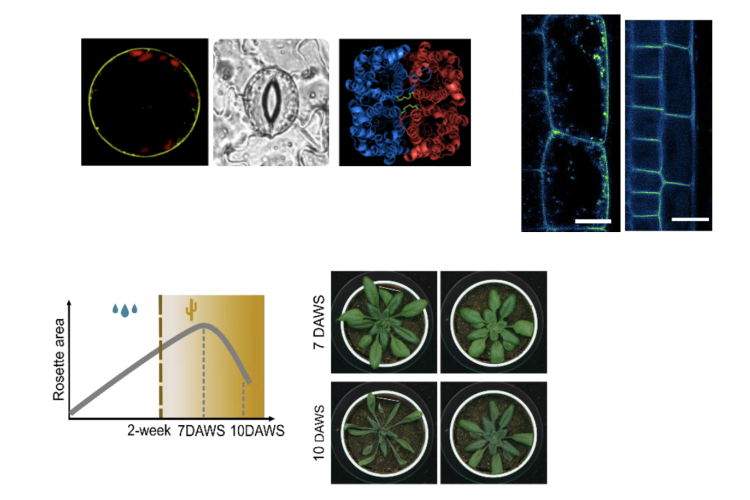
Interplay between the SCAMPs and PIPs in Arabidopsis
In eukaryotic cells, a network of organelles and membranes forms a highly organized and dynamic endomembrane system. The precise trafficking of plasma membrane (PM) proteins within this system is crucial for their proper function, ultimately influencing plant growth and development.
available scripts for image quantification
MitTally (Mitochondria Tally marks)
WIDEFIELD LIGHT MICROSCOPY
Under construction
Microscopy Imaging tools at PSB
Microscopy Imaging tools at PSB
PSB researchers are actively involved in generating tools to allow real-time visualization of plants under conditions that disturb specific processes. Examples are the use of microfluidics setups to monitor the effect of chemical compounds as well as the effect of altered temperature, on cellular dynamics.
Temperature-modulated live cell imaging
Temperature-modulated live cell imaging.
Dynamic imaging of fast-occurring processes is hampered by the fact that the signal/noise ratio of a fluorescent image is inversely related to the exposure time. The exposure time in turn limits the imaging speed and thereby the temporal resolution. We are increasing the temporal resolution of live-cell imaging plant samples by modulating the temperature of the samples in situ on the microscope stage as lowering the temperature slows down biological processes.
Developing proximity biotinylation assays in plants to expand the interactomics toolbox
Developing proximity biotinylation assays in plants to expand the interactomics toolbox.
Proximity biotinylation uses a promiscuous biotin ligase, which causes biotinylation of proteins in the vicinity of the bait. These biotinylated proteins can be identified using mass spectrometry without the need to maintain the protein-protein interactions during the purification. This tool is therefore especially suited for interactions between cytosolic and transmembrane proteins. We are designing protocols to adequately perform proximity biotinylation in plants.
Visualizing protein-protein interactions in plants
Visualizing protein-protein interactions in plants
To gain insight into protein-protein interactions that occur in planta, we expanded the interaction assay toolkit. We have developed Knocksideways in plants (KSP) as an imaging-based protein-protein interaction assays to visualize binary as well as higher-order interactions by. KSP can be compared to an intracellular Co-IP experiment. The tool uses the ability of rapamycin to alter the localization of a bait protein and its interactors via the heterodimerization of FKBP and FRB domains.
Stereomicroscopes
STEREOMICROSCOPES
Macroscope MVX10 Olympus